The NoisyR manuscript was accepted to Nucleic Acids Research (June 2021)!!
noisyR: Enhancing biological signal in sequencing datasets by characterising random technical noise
I. Moutsopoulos, L. Maischak, E. Lauzikaite, S. A. Vasquez Urbina, E. C. Williams, H. G. Drost, I. Mohorianu
https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkab433/629...
High-throughput sequencing enables an unprecedented resolution in transcript quantification, at the cost of magnifying the impact of technical noise. The consistent reduction of random background noise to capture functionally meaningful biological signals is still challenging. Intrinsic sequencing variability introducing low-level expression variations can obscure patterns in downstream analyses.
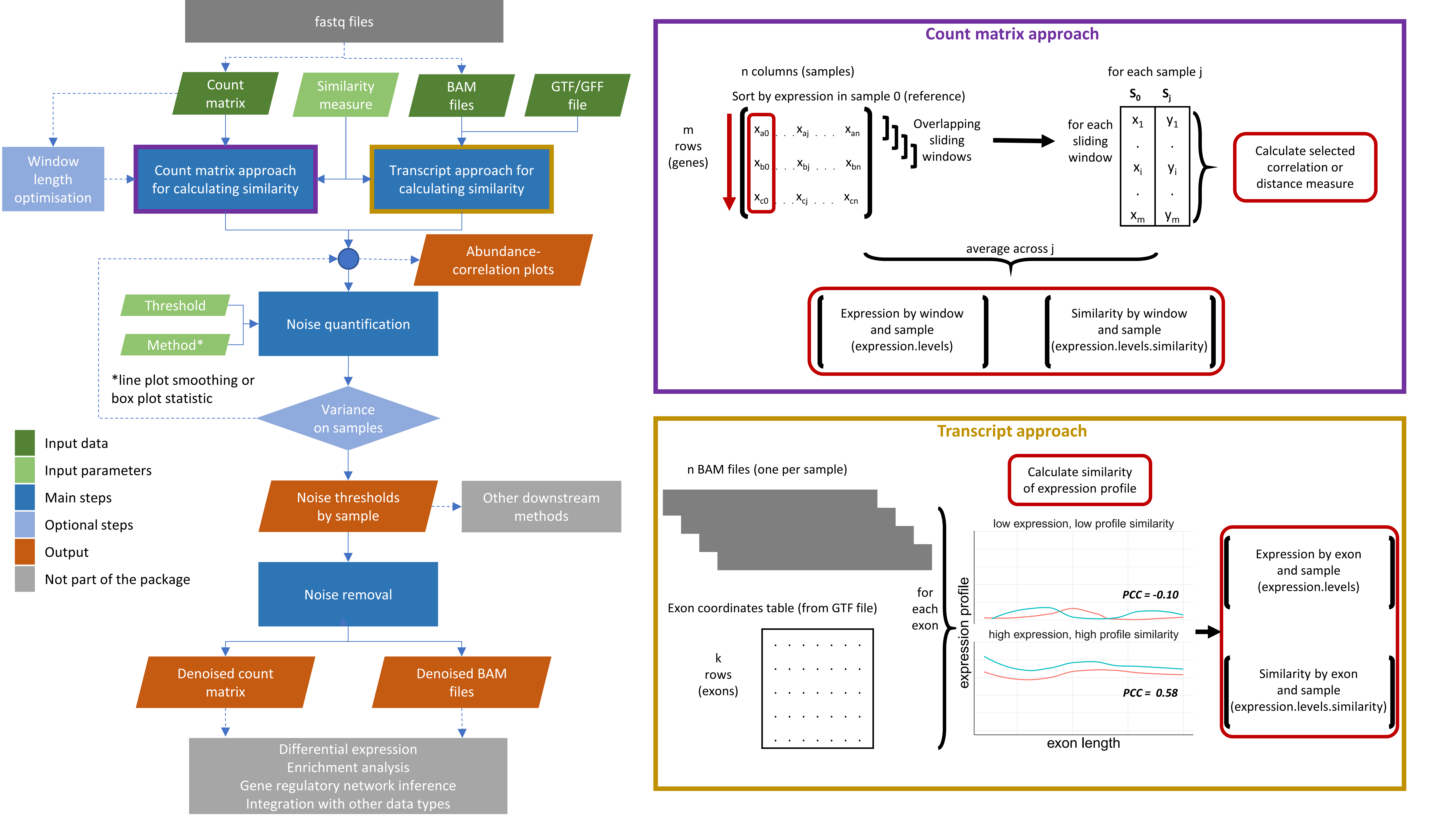
We introduce the noisyR package, an end-to-end pipeline for quantifying and removing technical noise from HTS datasets. The three main pipeline steps are [i] similarity calculation across samples, [ii] noise quantification, and [iii] noise removal; each step can be finely tuned using hyperparameters; optimal, data-driven values for these parameters are also determined.
Manuscript preprint: https://www.biorxiv.org/content/10.1101/2021.01.17.427026v2
Github page: https://github.com/Core-Bioinformatics/noisyR
Documentation: https://core-bioinformatics.github.io/noisyR/
Workflow diagram of the noisyR pipeline
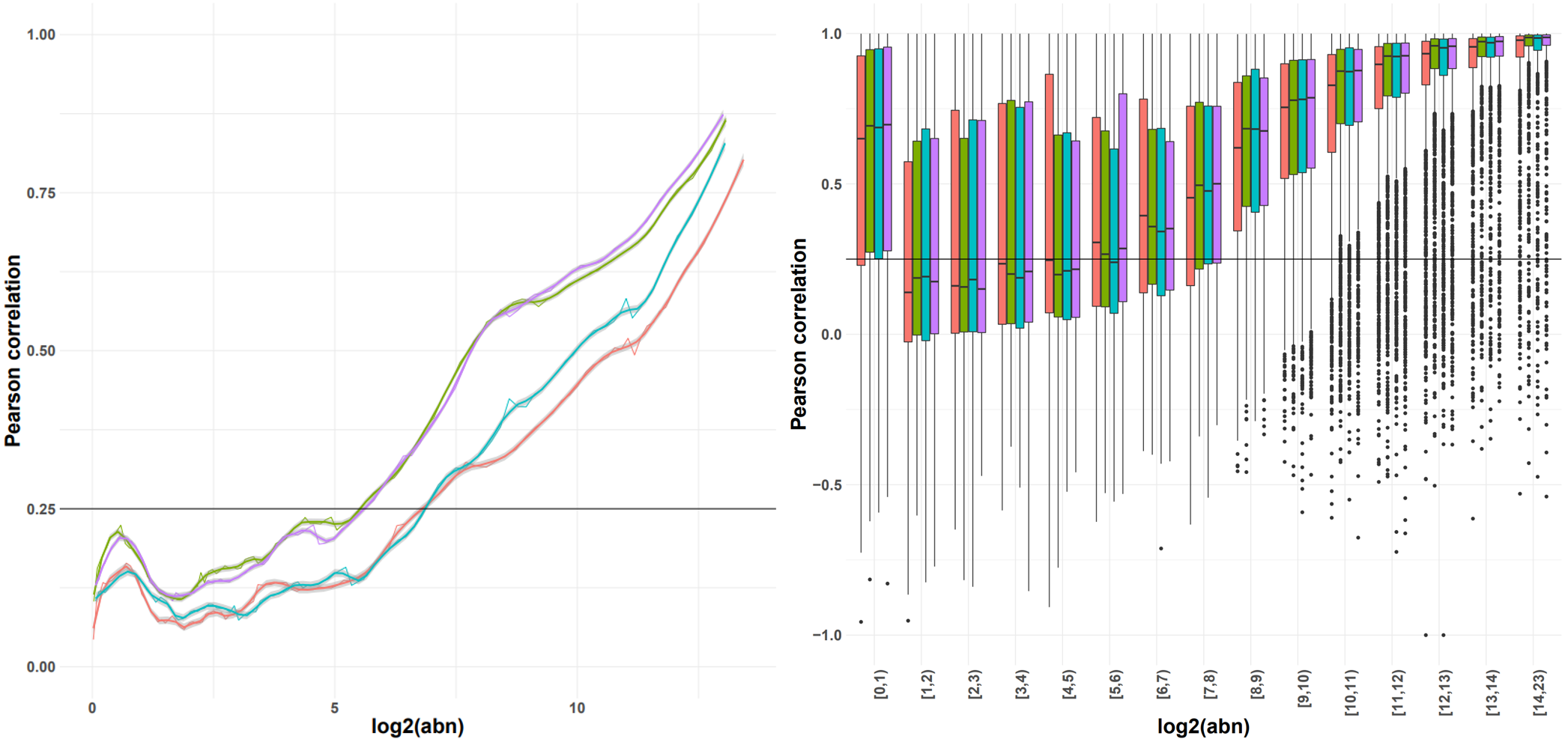
Indicative plots of the Pearson correlation calculated on windows of increasing average abundance for the count matrix-based noise removal approach (left) and per exon for the transcript-based noise removal approach (right).