For the optimisation of FACS-sorted cell cultures, we have generated a pipeline identifying discriminative genes from index sort data. The pipeline uses the depth distribution of features in the trees making up random forests.
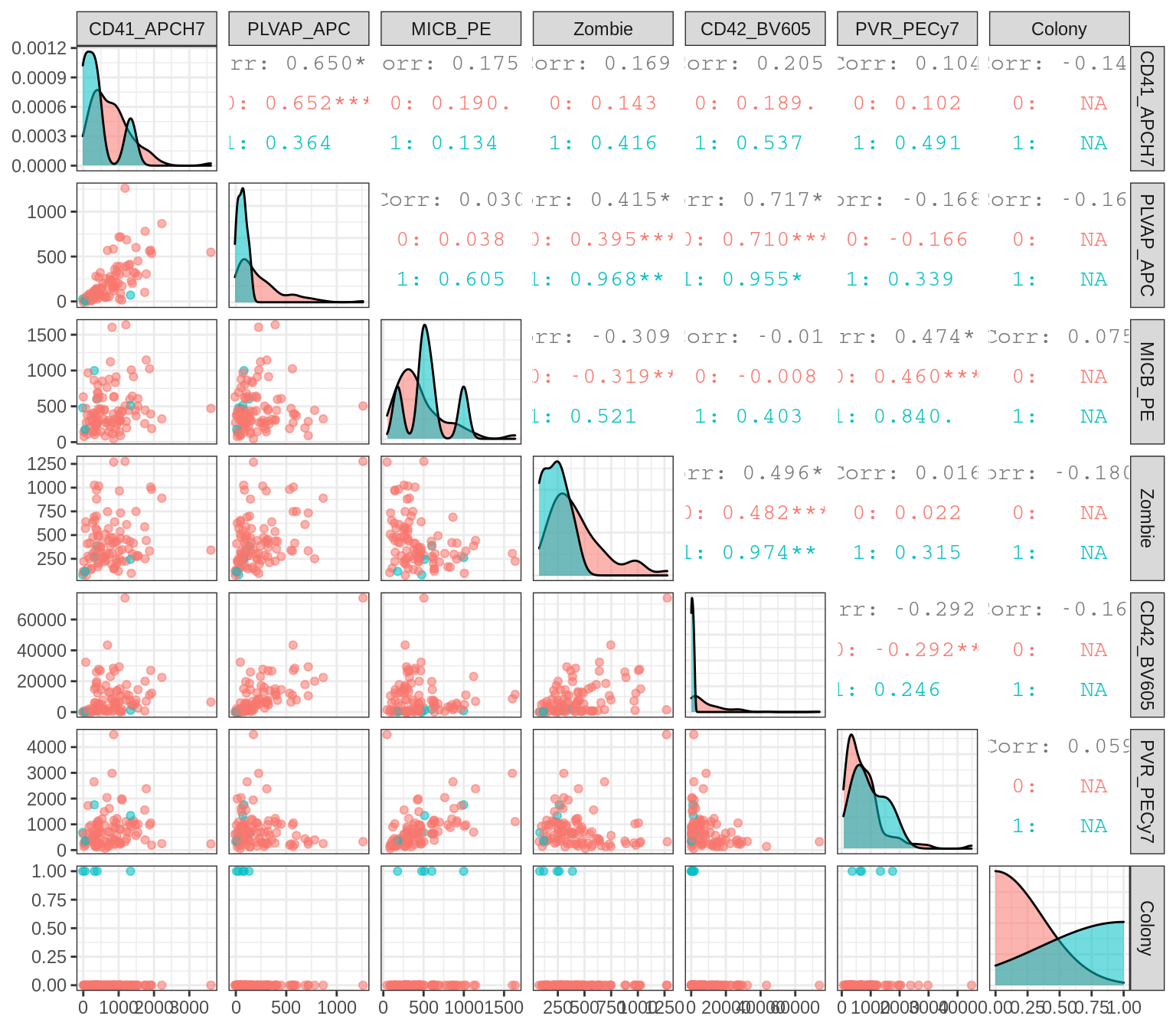
Densities and pairwise scatterplots for index sort features.
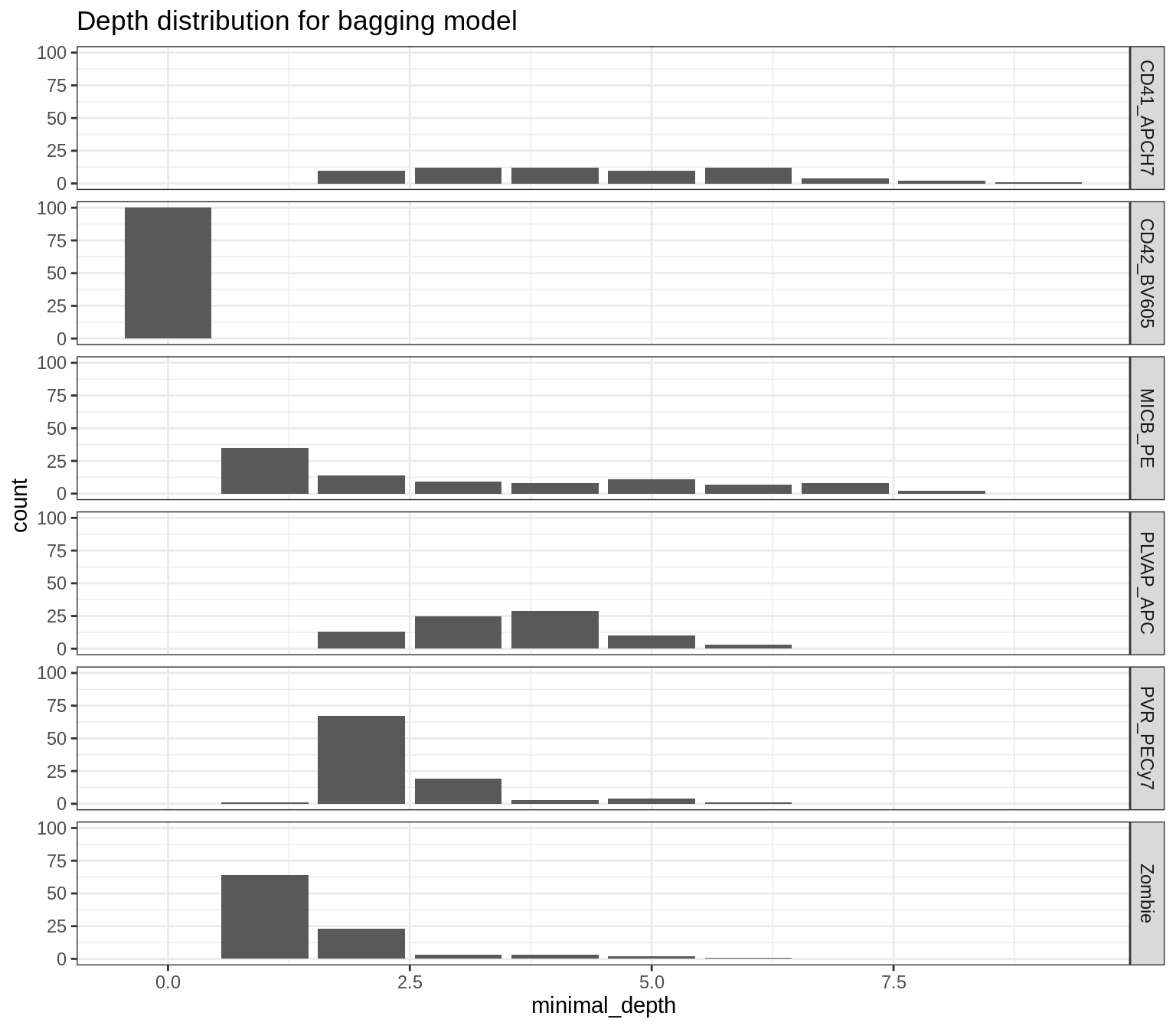
Depth distributions of features helps us identify the discriminative features (those with lower depth).
Github: https://github.com/Core-Bioinformatics/Lawrence-et-al-2021-index-sort-fe...
Manuscript: Mapping the biogenesis of forward programmed megakaryocytes from induced pluripotent stem cells by Moyra Lawrence *, Arash Shahsavari *, Susanne Bornelöv *, Thomas Moreau, Katarzyna Kania, Maike Paramor, Rebecca McDonald, James Baye, Marion Perrin, Maike Steindel, Paula Jimenez-Gomez, Christopher Penfold, Irina Mohorianu, and Cedric Ghevaert