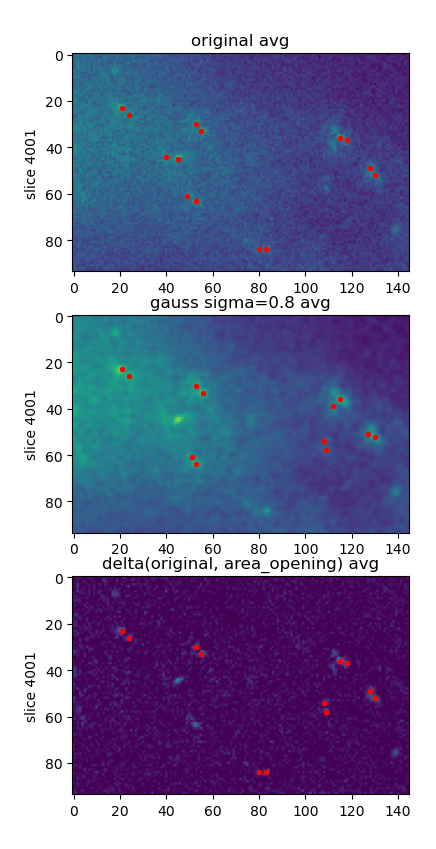
We are developing an imaging processing pipeline which would help us achieve the 3D localization of the fluorescent molecules analysed with the DHPSF microscope. The video is first stacked using a sliding window approach, and then we perform a processing over the resulting window. Through this method we obtain another processed video. The main challenges we encountered are the noisy datasets containing the background light emission from intracellular structures, moving structures, computational time restrictions and limited human interaction. For filtering the correct set of beads we are combining the processings using a voting scheme.