The transition from bulk to single-cell analyses transformed the computational challenges for high-throughput sequencing data processing. At the core of single-cell pipelines lies the partitioning of cells into subpopulations and characterizing the subpopulations. Given the extensive consequences that derive from this step, generating robust and reproducible outputs is essential.
Here we present ClustAssess, a suite of tools for quantifying clustering robustness both within and across methods. The tools provide fine-grained information enabling (a) the detection of the optimal number of clusters, (b) identification of regions of similarity (and divergence) across methods, and (c) stability-based evaluation of parameters in the common nearest neighbor-graph partitioning pipeline for clustering single-cell data.
The aim is to aid practitioners in evaluating clustering methods and parameters by providing information on how robustly cell identities are inferred from the resulting clusterings of their data.
Preprint:
Github: https://github.com/Core-Bioinformatics/ClustAssess
Documentation: https://core-bioinformatics.github.io/ClustAssess/index.html
CRAN: https://cran.r-project.org/package=ClustAssess
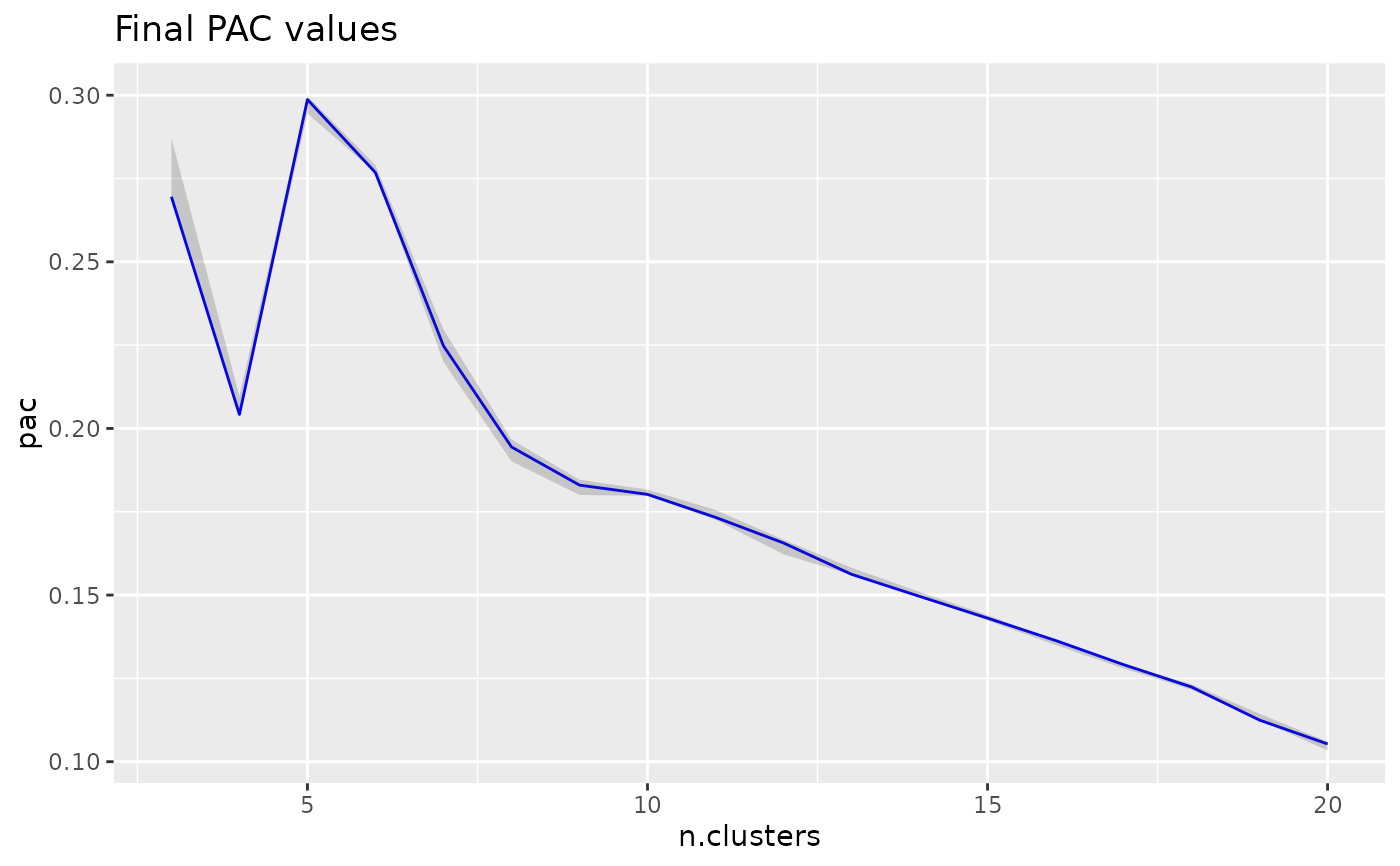
The optimal number of clusters can be determined using proportion of ambiguously clustered pairs (PAC).
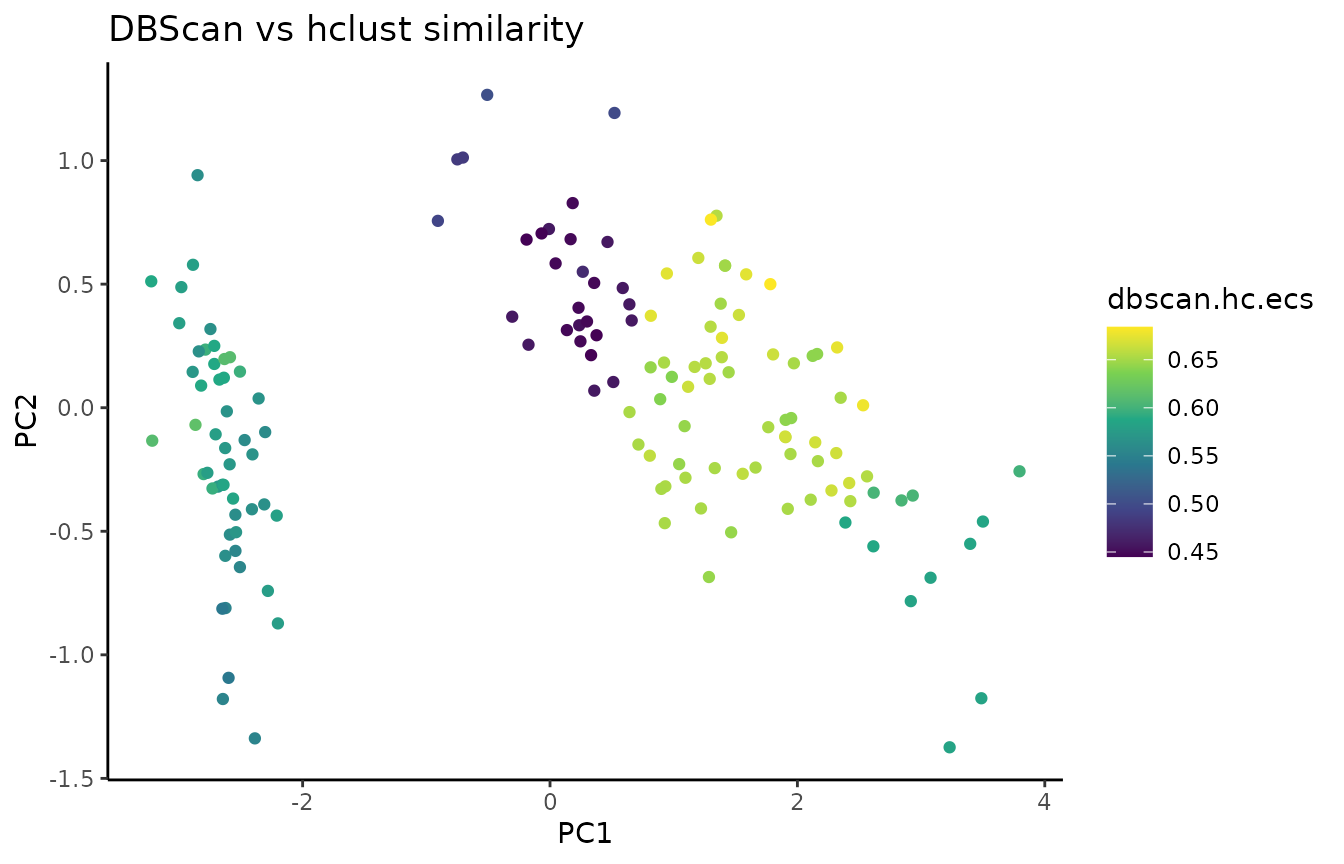
Element-centric clustering similarity (ECS) quantifies per-cell clustering agreement across methods.


